By Natalie Losada
The COVID-19 pandemic began a wave of virtual workshops that will ripple into the future because of their convenience and global attendance. However, some things are still better taught in person, like iJOBS Software Carpentry workshop to learn about Shell (short computer instructions), GitHub (file version tracking and storage), and R (language for long computer instructions). Shell should be learned for any computational study, while Git can be used for any file system you need to be tracked and shared. And R, as explained below, is niche to biostatistics.
This two-day workshop was possible because of the amazing instructors from both the east and the west coast. We had Heather Ciallella (she/her) from Rutgers University-Camden and Luna L. Sanchez Reyes (she/her)from University of California, Merced, who joined us over Zoom. Our helpers for the sessions were Kate Douthat (she/her), Jiayi Liu (she/her), and Kellen Xu (pronouns not shared) from Rutgers.
History
Software Carpentry is part of a non-profit organization that has existed since 1998. After merging with Data Carpentry, they became a single, non-profit incorporated called The Carpentries. They aim to teach researchers the computing skills necessary for working more efficiently with their files or raw data. They have volunteer instructors that have, since 2012, run hundreds of events for researchers all around the globe using their freely available lesson materials.
Last year The Carpentries workshop offered in partnership with iJOBS was a Genomics Data Carpentry workshop in a completely virtual format. So, keep an eye out for other Carpentry workshop themes in the years to come.
Lessons from the Workshop
Our instructor’s goal on the first day was to help us become comfortable with navigating our computers using a terminal emulator, which is a simple interface to talk to the computer one command at a time. If you’re imagining a computer hacker typing into a black screen with white text rapidly flying by as they typed, then you’re imagining the correct thing. The only difference is we were moving much slower. Mac users had Terminal already on their computer that could understand Shell commands we were learning. Windows users had to install the Git software that comes with the terminal emulator Git Bash. We needed these emulators because they use Shell to translate our commands to the brain of the computer. This is something we needed to know existed, but we didn’t need to know the inner workings of Shell or the terminal. The main reason for learning how to speak to the computer is to be able to automate tasks in high throughput.
For example, if you study structural biology like me, you might be trying to compare structures of different enzymatic proteins, i.e. tiny cellular machines. You might be working with dozens of files containing 3D protein coordinates, each containing many thousands of atoms. Additionally, each protein is surrounded by hundreds of water molecules. So, the volume of data is quite large. Finding anything in all the files would require a couple commands, or even a script, that checks each file and makes a nice list. Point being, once you know how to talk to the computer, you can let it do all the hard work for you.
Because this workshop was geared towards researchers, and researchers generate and analyze a lot of data, the second day of the workshop focused on file “version control” using Git locally (on your computer) and GitHub in the browser. GitHub might sound more familiar if you’ve needed to reference or share code publicly when publishing a paper.
Git is a system – free and open source – that keeps track of all versions of files and folders, even when multiple people are applying changes. For example, consider the editorial process between a lab member and a principal investigator (PI). Keeping track of newer and newer versions of the document can become wearying and overwhelming. Ideally the “track changes” setting in Microsoft Word is turned ‘on’ so tracking edits is facilitated. If you’re writing code, however, you’re not using Microsoft Word to track the changes, which is why many coders use Git to keep track of their updated files. Git is a great way to keep track of any file type, especially if multiple people are editing them (see image below), which is why it is called a “version control system”.
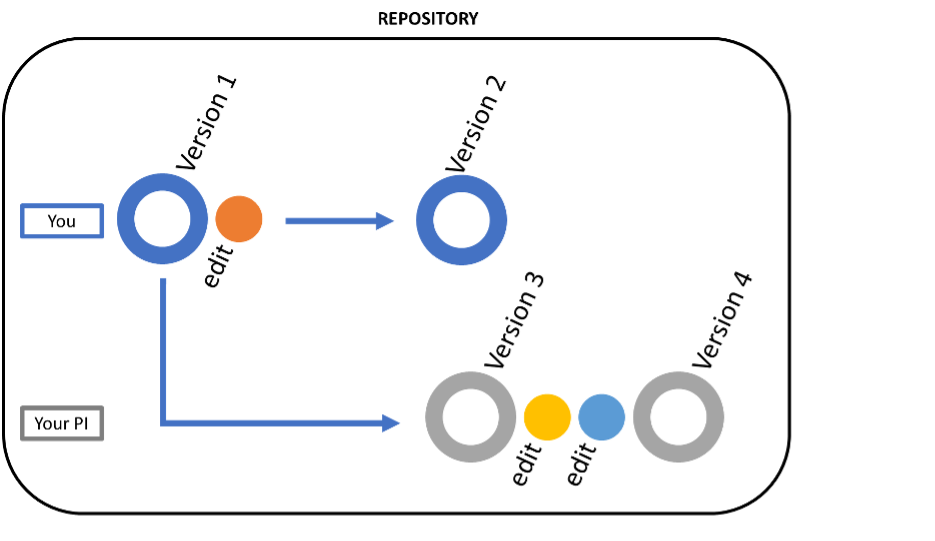
As shown in the above image, different file versions, as well as the specific changes to the file, are kept in the same repository. All you need to do is setup your Git account and tell it which folder to track, and it will follow all changes to the subfolders as well. The actual list of commands to set up Git is more involved, so check out the links at the carpentry website to set it up for yourself.
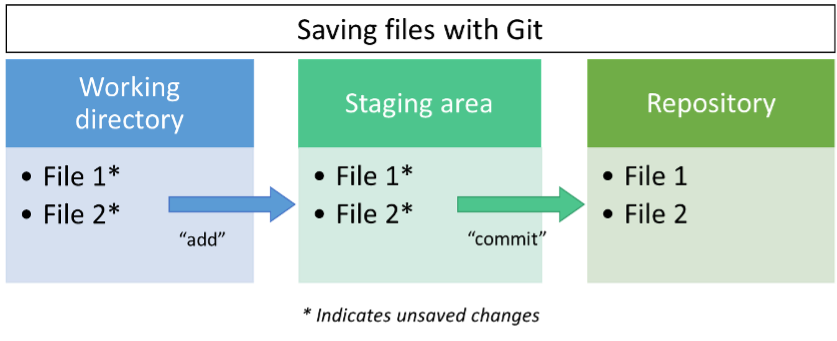
Once Git was ready for us to use, we had to understand one basic, but crucial concept of saving files to the Git repository. This can best be explained using the image below. As you change the files in your working directory (the specific location on your computer that Git is tracking) you are creating unsaved changes. When you have made enough progress, instead of hitting a save button, you go to the terminal and use the command “add” to send the unsaved changes to the staging area. This is followed by using the “commit” command to save the unsaved changes from the staging area to the Git repository. If you’re like me, this seems like a waste of time to use two commands to save a file. However, the “commit” command can work on multiple files at one time. At the end of the day, once your 22 (or however many) files have been updated with edits, you can “commit” them all at once to your repository and shut off your computer in peace knowing that your work is safe and sound. And because Git is linked to your account, you can login to GitHub anytime to view your work and share it with the public, coworkers, or keep it private.
During the afternoon sessions of both days, we focused on learning a computer language called R. Most of us had never used R before, so learning this language seemed like a daunting task. However, our virtual teacher, Luna, had entirely taught herself to write in R and was now passing on the knowledge.
For context, Shell is used to translate our commands to the computer via the terminal, but computer languages translate via their application environment or script files that can be referenced in Shell. If you have not heard of the language R, you may have heard of Python. Python is a relatively easy-to-learn language for programming and has a lot of versatility in what it can be used for. It is closer to human-readable English than some other languages designed and released in previous decades. R is also easy-to-learn and, arguably, similar to Python. However, compared to Python, R isn’t versatile in function. On the other hand, R does perform its main objective very well, which is statistical computing and making graphical data representations.
Luna led us through the basics of the language to get us comfortable with the syntax and the RStudio software that facilitates writing scripts in R. We learned commands to manipulate files, create variables, and use functions. Now, what on earth does that mean? When using R, you need to separately tell it to open files to be able to look at the contents. Additionally, when asking the computer for information from the file, you need to be precise. You cannot ask the computer to look at file.txt because it won’t know where to look. You need to say where the file is saved as well. Computers are smart, but they can’t infer what you were trying to say. After that we learned how to take those files and pieces of files and turn them into variables for functions. If you recall your algebra knowledge, you can remember that a variable, like x, holds the place of some information in an equation, or the function. When coding, the variable acts the same way, but you get to choose what x is! You can also customize functions or reference a function the computer already knows exists in RStudio. Functions can be tricky because of their versatility, but just think of them as instructions. If I told you to drink some water, you would know to find a glass of water, or if necessary, put water in a glass to drink the water. That whole flow of conditional actions could be written as a function while coding. And I can easily tell you to perform the function again with a different drink like soda. Computers work the same way.
The last thing we learned in RStudio was how to make graphs and plots. Explaining it in words is quite boring but making them is fun! So, I highly recommend taking a look at The Carpentries materials so you can explore and make your own figures.
Learning about Shell, GitHub, and R might have seemed like a lot of material for one workshop. But they are all essential in working with large amounts of data quickly and efficiently, as us researchers need to do sometimes. Shell allows you use simple commands for file management that is then saved securely with GitHub, and R allows you to group longer, complicated commands into scripts than can be run and customized later. Together they are a powerful triad!
This article was edited by Junior Editor Shawn Rumrill and Samantha Avina.